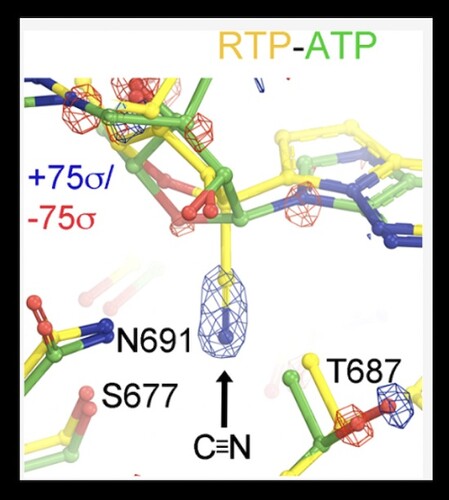
Remdesivir, the nucleotide analog, is known to interact with the SARS-CoV-2 nsp12 RNA dependent RNA polymerase (RdRp) and act via a delayed inhibition mechanism . While multiple Cryo-EM structures have been determined for Remdesivir within the replication-transcription complex (RTC) , there is a limited amount of detail that can be interpreted from these structures. In their new paper published in the Biochemistry journal of ACS publications, Wang and colleagues utilize molecular dynamics (MD) simulations to provide new insight into the binding mode and reaction mechanism of Remdesivir triphosphate (RTP). They first report on the nucleotide selectivity of RTP over ATP, as well as analyze detailed hydrogen bond networks that are difficult to resolve in experimental maps. Modeling the differences in the binding pocket with ATP versus RTP allowed the researchers to propose hypothetical models with other chemical moieties, which may inform future drug design to outperform Remdesivir. The study is a prime example of how MD simulations can provide the basis for structure-based drug design.
Authors affiliated with Yale University include Jimin Wang, Yuanjun Shi, Krystle Reiss, Federica Maschietto, Elias Lolis, William H. Konigsberg, and Victor S. Batista, with their collaborator George Lisi from Brown University.
By Shravani Balaji