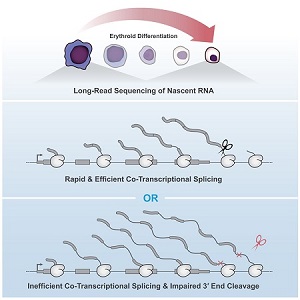
Kirsten Reimer, an MB&B graduate student in Karla Neugebauer’s group, recently published a study in Molecular Cell which found that co-transcriptional splicing of pre-RNA influences 3′ end cleavage efficiency. By investigating transcription elongation and 3′ end formation in differentiating murine erythroblasts, Reimer and her co-authors determined that intron retention was correlated with co-transcription splicing efficiency. This association was consistent within the same transcript.
Combining long read sequencing of nascent RNA with precision run-on sequencing (PRO-seq), Reimer was able to monitor co-transcriptional splicing during transcription and 3’end formation. Introns transcribed during erythropoiesis were frequently removed co-transcriptionally and in a coordinated process. Interestingly, despite no apparent pause in transcription following splicing, there were several intermediate splicing variants. This indicates that delays can occur between the catalytic steps of splicing.
According to Dr. Neugebauer, their unique method enables researchers to view the point at which RNA polymerase and the spliceosome coordinate to generate nascent RNA molecules.
“Our method, long read sequencing of nascent RNA, allows us to see what RNA molecules look like as they are being born,” Dr. Neugebauer said. “[It] catches them in the act, telling us how the activities of the two machines are coordinated with one another. We found that human RNA polymerases and spliceosomes seem remarkably well coordinated, since the spliceosome is able to act on the RNA as soon as it is made.”
These results were surprising, considering the complexity of human genes and indications from other studies that there is limited coordination between synthesis and splicing. Yet there was also variability in the extent of coordination among certain genes examined by the researchers. One RNA in particular, globin mRNA, was significantly less well coordinated than other genes.
One mutation identified in a thalassemia patient created a cryptic splice site that improved splicing and 3’ end cleavage of beta-globin transcripts.
“To our surprise, even though the mRNA that is expressed from the mutant allele is eventually degraded by NMD, the pre-mRNA is spliced even more efficiently than the wild-type,” Reimer said.
The authors hope that their approach will enable future progress in continuing to decipher the coordination of RNA polymerase and the spliceosome, as well as the molecular bases for variable coordination among different transcripts.
Further details on the study and the full paper can be found in the January 12th issue of Molecular Cell.