Two new articles from Dr. Joan Steitz’s group published in Science and PNAS describe new RNA tertiary structure motifs, including a novel poly(A) binding pocket, and push the boundaries of small angle X-ray scattering (SAXS) and cryo-EM analyses on small RNA molecules. An article appearing this week in PNAS, by Seyed Fakhreddin Torabi et al., describes structural characterization of the interaction between two small RNA molecules – a double element for nuclear expression (dENE) and a 28-mer poly(A) – by SAXS, cryo-EM, and biochemical structure probing. This article builds upon their publication in Science last month, in which Torabi and colleagues determined the high-resolution structure of the dENE+poly(A) complex by X-ray crystallography.
Elements for nuclear expression (ENEs), located upstream of poly(A) tails of mRNAs or lncRNAs, slow the decay of RNA by forming a triple helix with the poly(A) tail. This stabilizing interaction is highly conserved and may have been important in early evolution because it can form without RNA-binding proteins. The results from these recent papers suggest that there may be several mechanisms by which ENEs can prevent RNA decay.
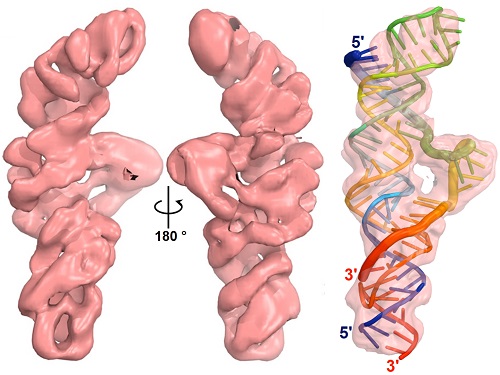
In their X-ray crystallography study published last month, the authors were surprised to find a molecular binding pocket in the interaction between the poly(A) and the dENE. “We knew that this RNA element was different from the previous triple helix structures and should have some new structural features, however we were not expecting to observe a molecular RNA pocket” said Torabi. The pocket hides the extreme 3′ end of the poly(A) tail and protects RNA from 3′-end degradation through a steric mechanism.
In their article coming out this week in PNAS, Torabi and colleagues in the Steitz Lab, the Pollack Lab at Cornell and the Chiu Lab at Stanford used biochemical assays, SAXS and cryo-EM to validate that the structure obtained by X-ray crystallography was also valid in solution. The structures presented in this paper are some of the smallest macromolecules to be determined at sub-nanometer resolution using cryo-EM. The authors were also able to observe local conformational changes in the dENE upon complex formation with poly(A). Furthermore, they demonstrated that, by extending the RNA or incorporation of bulky groups, the orientation of the molecule and directionality of the binding of another RNA to a small RNA element can be addressed by SAXS molecular reconstructions.
The work by Torabi et al. presented in these two papers introduces new RNA tertiary structures, expands our knowledge of the role of RNA-poly(A) interactions in RNA stabilization, and contributes to an understanding of the evolution of poly(A) tails on mRNAs.
The full articles can be found here:
Science, February 5, 2021 issue
By Melanie Reschke