A grand challenge of modern cryo-electron microscopy (cryo-EM) is to determine high-resolution biological structures in their native states in the subcellular context. Structural studies on the very large, flexible, and highly dynamic cytoskeletal motors bound to their intricate tracks in an exceedingly crowded environment are such examples. For decades, these have been studied at very low resolution when the targets cannot be defined as ‘dispersed particles’ in solution for conventional single-particle cryo-EM.
Dynein is a microtubule-based cytoskeletal motor which drives intracellular cargo transport, dynamics of many cellular materials and ciliary beating. High-resolution structures of dynein bound to microtubules have been desired for decades, as they are critical to advancing the mechanistic understanding of dynein’s cellular roles in the field. Despite the recent resolution revolution in the realm of single-particle cryo-EM targeting protein complexes as soluble, dispersed particles in solution, structure determination of dynein complexes irregularly bound to the highly curved, intersectional microtubules remains a major challenge. Traditionally, solving high-resolution structures of microtubule-associated proteins (MAP) bound to microtubules requires pre-reconstruction of the microtubule itself and demands several strict conditions like dense, regular decoration of the MAP. However, the large size and highly dynamic nature of dynein complexes defy these rules, making standard cryo-EM image processing pipelines inapplicable.
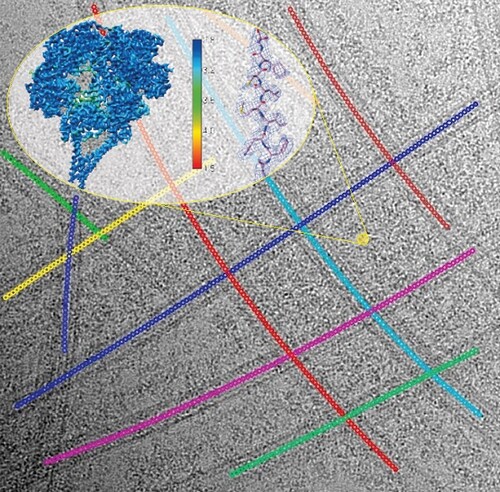
In a recent method paper published in Journal of Structural Biology, Pengxin Chai et al. devised a novel solution. They first trace the crowded microtubule networks by an iterative multi-curve fitting approach, which accurately determines the central coordinates of individual microtubule segments, reliably clusters the coordinates belonging to the same curve in the network, performs geometrical parameter optimization and eventually approximates the whole microtubule networks as a sect of analytical functions for subsequent image analysis. Based on the multi-curve fitting, they accurately estimate the principal signals of tubulin-lattice and computationally subtract the signals from raw cryo-EM micrographs. This novel method bypasses the standard microtubule reconstruction pipeline and allows direct 3D reconstruction of dyneins on microtubules at high-resolution. Using outer-arm dynein bound to microtubules as an example, the authors demonstrate the capability of the new approach and report the first high-resolution structures of dynein complexes bound to microtubules.
The authors also describe that the new method can be readily applied to other dynein complexes to advance our understanding of the intricate dynein molecules. In fact, the method immediately had positive impacts on high-resolution structures of highly challenging motors on the cytoskeleton. For example, two recent works from Carter’s lab have already adopted the method and reported near-atomic structures of dynein-dynactin-adaptor complexes bound to the microtubule. These excellent examples demonstrate the importance of methodology development that really drives a field forward.
In the future, the authors’ goal is to generalize the method to enable near-atomic structure determination of a myriad of biological complexes bound to cytoskeletal networks. The work was led by Pengxin Chai, Qinhui Rao, and Kai Zhang. The paper can be found in the Journal of Structural Biology.
By Kai Zhang