New work from MB&B graduate student, Max Paul, published in PNAS this week describes how a highly conserved region of the Ras GTPase-activating protein (GAP) C2 domain augments enzymatic activity through an allosteric interaction. Max, who is in the Boggon lab, studied the archetypal GAP protein, RasGAP, to assess the function of its C2 domain. While C2 domains are known to be a membrane-targeting calcium-binding domain, these traits are not necessary for this fold. It has been suggested in the field that C2 domains may play a role in GAP enzymatic activity, but to date, its function has yet to be rigorously examined.
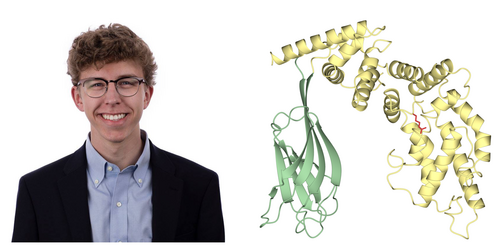
Paul and colleagues began by confirming that the C2 domain of RasGAP increases the catalytic activity of the adjacent GAP domain, comparing C2-GAP to GAP alone in in vitro multiple turnover GAP assays. With data to suggest the C2 domain is important for complete GAP activity, Max sought to visualize of the C2-GAP domain to understand its function. He determined a 2.45Å crystal structure of the C2-GAP domain region of RasGAP. When this C2-GAP domain structure was superposed onto previously determined structure of the GAP domain in complex with Ras, they found an intriguing proximity between the C2 domain and Ras, only 7 Å apart. Additionally, AlphaFold heterodimer predictions suggest a direct interaction between the C2 domain and the Ras allosteric lobe. Together, the structure and subsequent predictions suggest direct contact between RasGAP C2 and Ras. To further support their unexpected finding, they then aligned hundreds of sequences of RasGAPs from humans to sponges, and found the C2 domain surface closest to Ras is completely conserved.
The study then identified a residue, R707, that may be involved in the direct interaction with Ras, and asked whether mutation of this residue could affect RasGAP catalytic activity. Through a single turnover phosphate release assay, they found mutating R707 negated the gain that the C2 domain provided to GAP activity. To extend the implications of their findings, they sought to understand the in vivo effects of this mutation. In collaboration with the King lab at the University of Michigan, they used CRISPR/Cas9 targeting to generate a Rasa1 R698C allele in mice, equivalent to human RASA1 R707C. They found the mutation to be embryonic lethal in homozygous form; additionally, they found it impaired vascular development of surviving mice due to the catalytic inactivation of RasGAP and subsequent upregulation of the MAPK/ERK pathway.
These findings may extend beyond RasGAP. Upon sequence alignment and structural mapping of all other C2 domain-containing RasGAPs, Max found the residues to be almost identical, with R707 almost completely conserved. He then probed SynGAP, introducing an equivalent mutation, and found catalytic activity to be abrogated. Max is especially excited for this finding, as these results could indicate a previously underappreciated mechanism of Ras regulation common to almost all members of the RasGAP family, implicating the allosteric lobe of Ras as an important regulatory site.
Read more on this study here.
Shravani Balaji