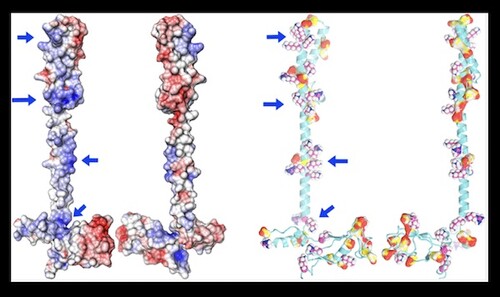
Jimin Wang and colleagues recently published in the Biochemistry journal of ACS Publications on “Insights into Binding of Single-Stranded Viral RNA Template to the Replication−Transcription Complex of SARS-CoV‐2 for the Priming Reaction from Molecular Dynamics Simulations.” In this study, researchers looked at the electrostatic potential distribution and carried out molecular dynamics simulations of the minimal replication-transcription complex (RTC), composed of the nsp12 RNA-dependent RNA polymerase, two nsp8 primases and one accessory subunit nsp7.
The priming reaction, composed of primer synthesis and primer elongation, is an essential step of RNA replication and transcription. However, there remains a gap in knowledge about how the template binds at the RTC and becomes competent for priming.
In the original SARS-CoV, nsp7 and nsp8 are thought to form a stable hexadecamer with a nucleic acid-binding central pore. This mechanism has not been observed with SARS-CoV-2 within the RTC. Molecular dynamics (MD) simulations revealed that optimal binding for the active priming complex most likely requires one nsp8 in the nsp7-bound conformation on one side of the template, and a free nsp8 on the other side. The distribution of electrostatic potential maps obtained from these MD simulations are in agreement with multiple cryo-EM maps of this complex. Researchers explored whether the conformations of the nsps would vary when considering the primer/template duplex or an ssRNA template, but found it likely that the conformations may resemble each other. Overall, the paper provides valuable insight into the initial reactions required for the synthesis of viral RNAs.
Authors affiliated with Yale University include Jimin Wang, Yuanjun Shi, Krystle Reiss, Brandon Allen, Federica Maschietto, Elias Lolis, William H. Konigsberg, and Victor S. Batista.
By Shravani Balaji