Conformational Landscape of Reactive Dynein-1 Unveiled by Dynamic Cryo-EM Analysis
A collaborative study, led by the Zhang Lab at Yale University and the Steven Markus lab at Colorado State University, unveiled the most complete conformational landscape of the reactive dynein-1 in atomic detail, offering unprecedented insights into how this massive molecular machine powers essential intracellular transport.
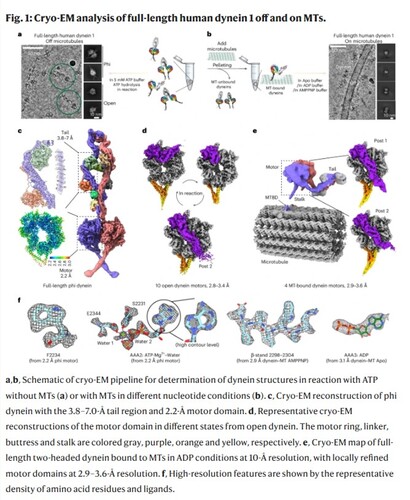
Published in Nature Structural & Molecular Biology in a study titled “The Mechanochemical Cycle of Reactive Full-length Human Dynein-1”, the research presents over 40 high-resolution cryo-electron microscopy (cryo-EM) structures of full-length human dynein-1 in a variety of nucleotide and microtubule-binding states. The resulting conformational landscape provides a dynamic view of how dynein converts chemical energy from ATP into mechanical motion — a process known as the mechanochemical cycle.
“Dynein is incredibly complex, and until now, we’ve only had fragmented glimpses of how it works due to the limited number of conformational states previously available,” said Pengxin Chai, co-first author and recent Ph.D. graduate in the Zhang Lab at Yale’s Department of Molecular Biophysics and Biochemistry. Jun Yang, also co-first author and postdoctoral researcher in the same lab, added, “This study brings us closer to a full structural and mechanistic understanding of dynein in motion.”
Unlike previous structural studies that aimed to trap dynein in specific conformational states using nucleotide analogs, this study directly imaged reactive dynein in the presence of ATP, capturing the motor in action as it progresses through its full mechanochemical cycle. Using advanced heterogeneity analysis of cryo-EM, the researchers identified numerous novel intermediates and revealed a previously unrecognized “molecular backdoor” mechanism for phosphate release during ATP hydrolysis.
A major challenge in probing dynein’s cycle has been the lack of microtubule (MT)-bound dynein structures. By employing a cryo-EM method (Chai et al., 2022, JSB) previously developed by the Zhang Lab, the team captured high-resolution structures of dynein bound to microtubules. This enabled a comprehensive view of how dynein transitions between states on and off the microtubule, culminating in a revised model of dynein’s stepping and powerstroke mechanism — offering new insights into how dynein generates directional force during intracellular transport.
In addition to Pengxin Chai, Jun Yang, and Yue Wang from the Zhang Lab, co-author Indigo C. Geohring from the Markus Lab contributed key functional analyses, supporting the revised mechanistic models. This interdisciplinary effort exemplifies the power of frontier imaging technologies and highlights the importance of collaboration across structural biology, biochemistry, and computational biology, pushing the boundaries of our understanding of molecular motors.
The full paper is available in Nature Structural & Molecular Biology here.