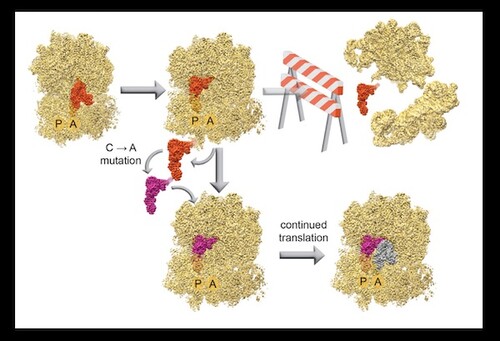
Genetic recoding is a powerful tool to utilize non-canonical amino acids and create highly diverse proteins. Researchers in MB&B have been working on expanding the genetic code through recoding, and recently published a biophysical study on a uniquely structured tRNA: allo-tRNAUtu1, that has been utilized for selenocysteine insertion at a stop codon sequence. Using single molecule fluorescence studies, they observed the progression of translation in real-time, tracking how this tRNA compared with a natural suppressor tRNA and establishing roadblocks associated with tRNAUTu1 translation.
Researchers then used Cryo-EM to visualize this synthetic tRNA on the E.coli ribosome; the resulting structure at 2.6Å allowed for atomic-level modeling and provided insight into distinct steps of tRNA movement along the ribosome. Their analysis led them to a distinct tertiary interaction that prevented fluid translocation of the tRNA. Through a single-base mutation in tRNAUTu1, they were able to relieve this barrier and enhance translation processivity on the ribosome, resulting in an increase of selenocysteine-incorporated protein production. Their work demonstrates the potential of synthetic tRNA substrates, especially when subject to biophysical analysis and subsequent engineering.
This study was led by Natalie Krahn and Oscar Vargas-Rodriguez and can be found in Nucleic Acids Research.
By Shravani Balaji