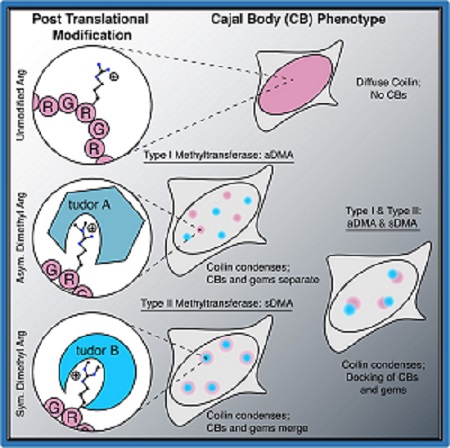
In an article published this week in Cell, the Neugebauer and Bewersdorf labs identify the interaction between protein tudor domains and dimethylarginine modified proteins as a driver and regulator of membraneless organelle assembly. Membraneless organelles include P-granules, Cajal bodies, and nucleoli and are formed by the condensation of proteins and RNA. The assembly and composition of these structures is highly specific, and understanding the mechanisms by which this assembly is regulated has been a recent focus of research in the field.
To investigate this, members of the Neugebauer and Bewersdorf labs studied the condensation promoting capability of different regions of the survival of motor neuron protein (SMN). SMN contains two intrinsically disordered regions (IDRs) and one tudor domain. Previous studies have pointed to IDRs in RNA binding proteins as drivers of condensation. However, in this study the authors unexpectedly found that the SMN tudor domain promoted condensation upon binding to dimethylarginine-modified protein ligands. The interaction of the tudor domain with different forms of dimethylarginine determined the composition and structure of the condensed membraneless organelles.
Edward Courchaine, Ph.D., the lead author of this study, commented on the group’s findings: “What’s most exciting to me is that we found a new way in which condensates can be regulated in a specific manner. We’ve known that the biophysical properties of proteins change as a result of arginine methylation, but the specific binding by Tudor domains could act more like a switch to control membraneless organelles. It’s a lot easier to imagine cellular events under that kind of control system, and our data shows that you can transition between separate versus co-condensation just by changing the methylation status.”
In a closing remark, Courchaine looked ahead to potential upcoming studies: “Going forward it will be fun to see which pathways are using this module to control assembly and what functional ramifications come out of that.”
The full article can be found here: https://www.cell.com/cell/fulltext/S0092-8674(21)00625-5
By: Melanie Reschke